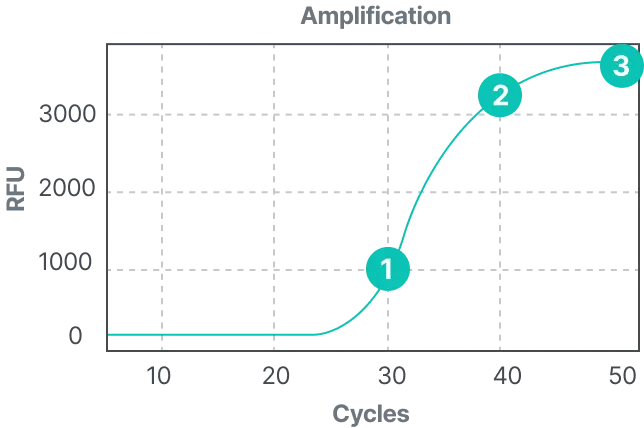
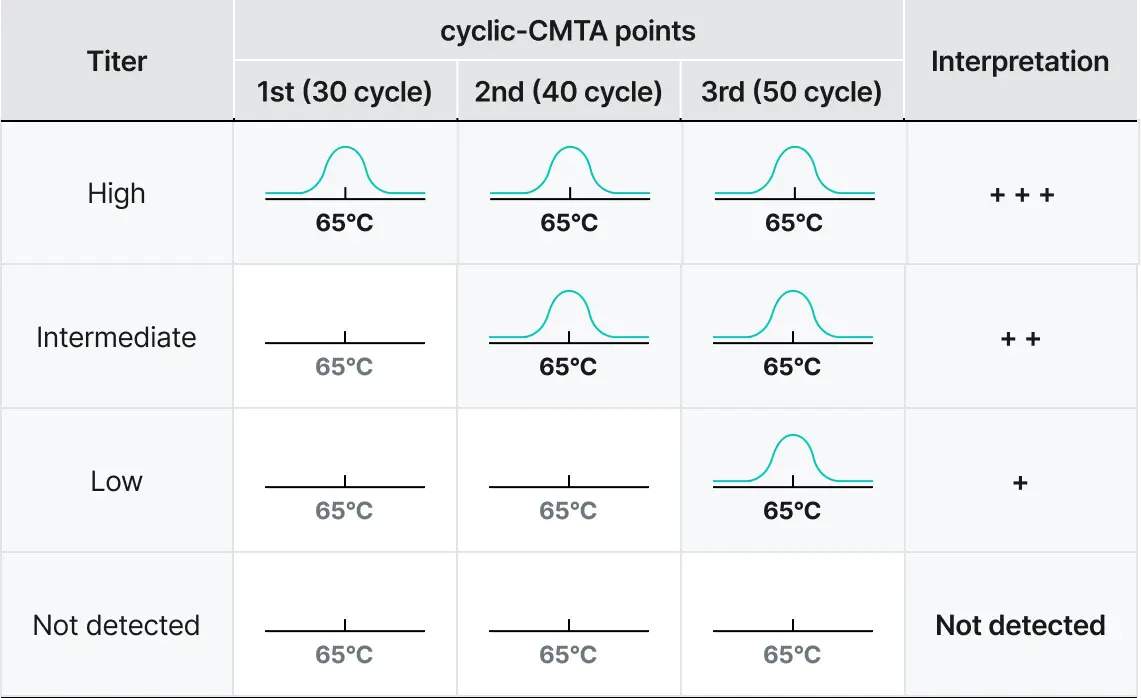
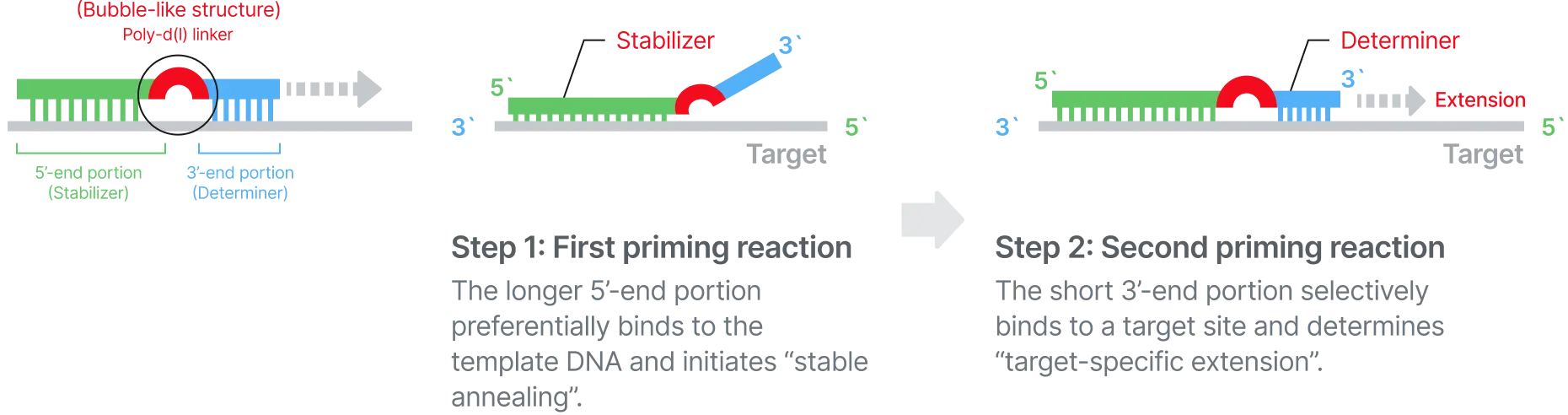
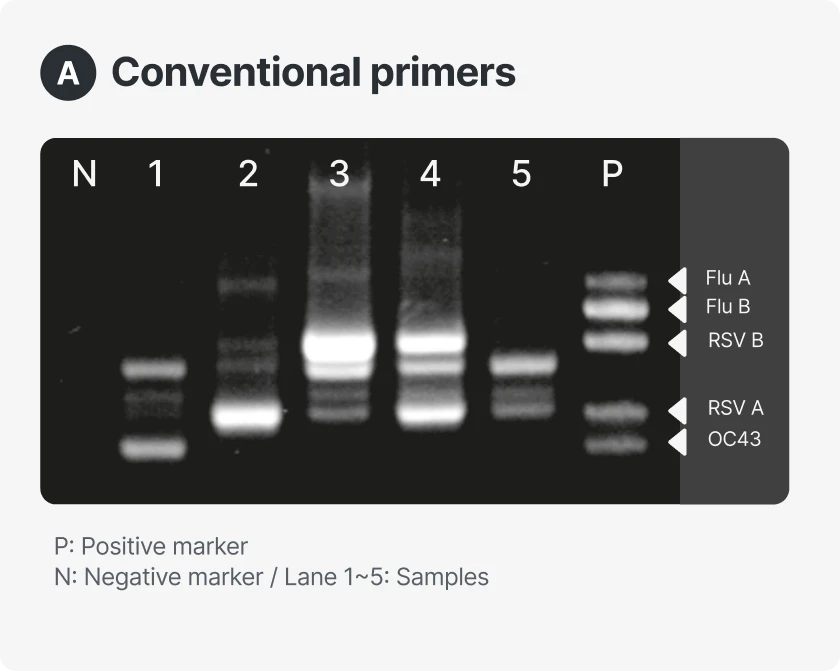
STST™ is a next-generation PCR reagent platform designed to generate independent, non-overlapping fluorescent signals at a single temperature. This structural signal separation eliminates the need for post-processing required in conventional multiplex PCR, enabling accurate quantification of up to five targets within a single channel. With a simplified analytical workflow and intuitive signal interpretation, STST™ improves both the speed and precision of diagnostics while maximizing multiplexing performance across various instrument environments.
STST™ Features
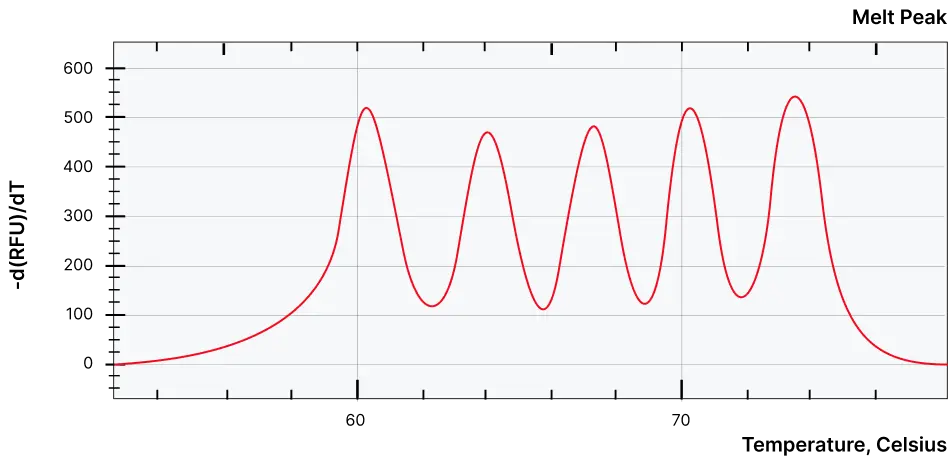
Quantitative multiplexing within a single channel
- Detects up to five targets in one channel
- Provides independent, non-overlapping signals without post-run processing
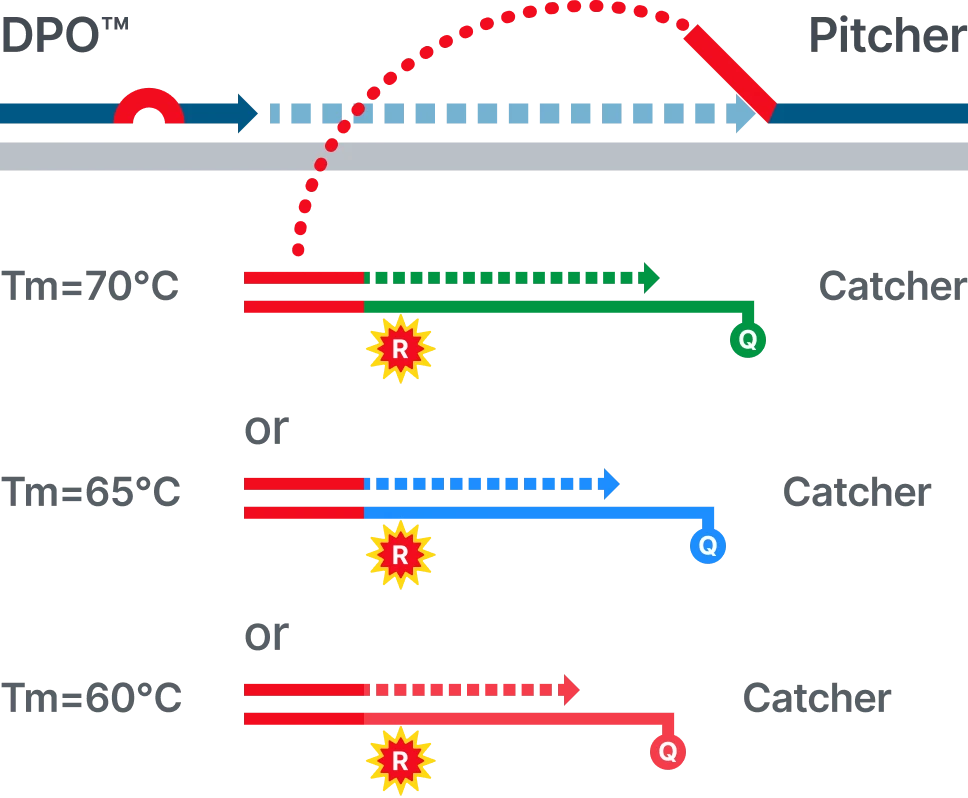
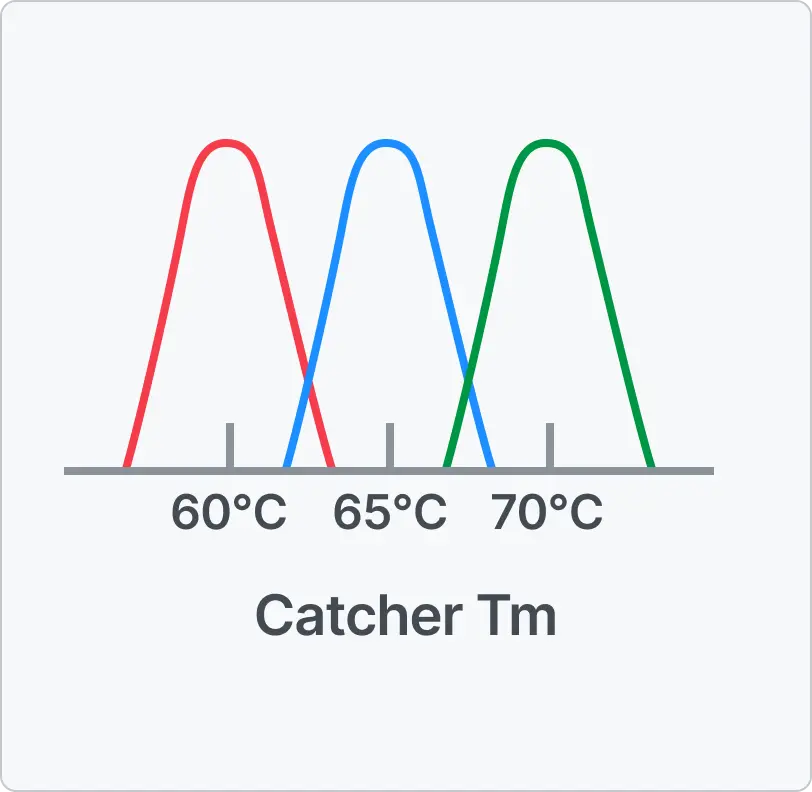
Direct and reliable signal interpretation
- “What you see is what it is” — stable signals directly reflecting each target
- No additional analysis or deconvolution required for Ct calculation
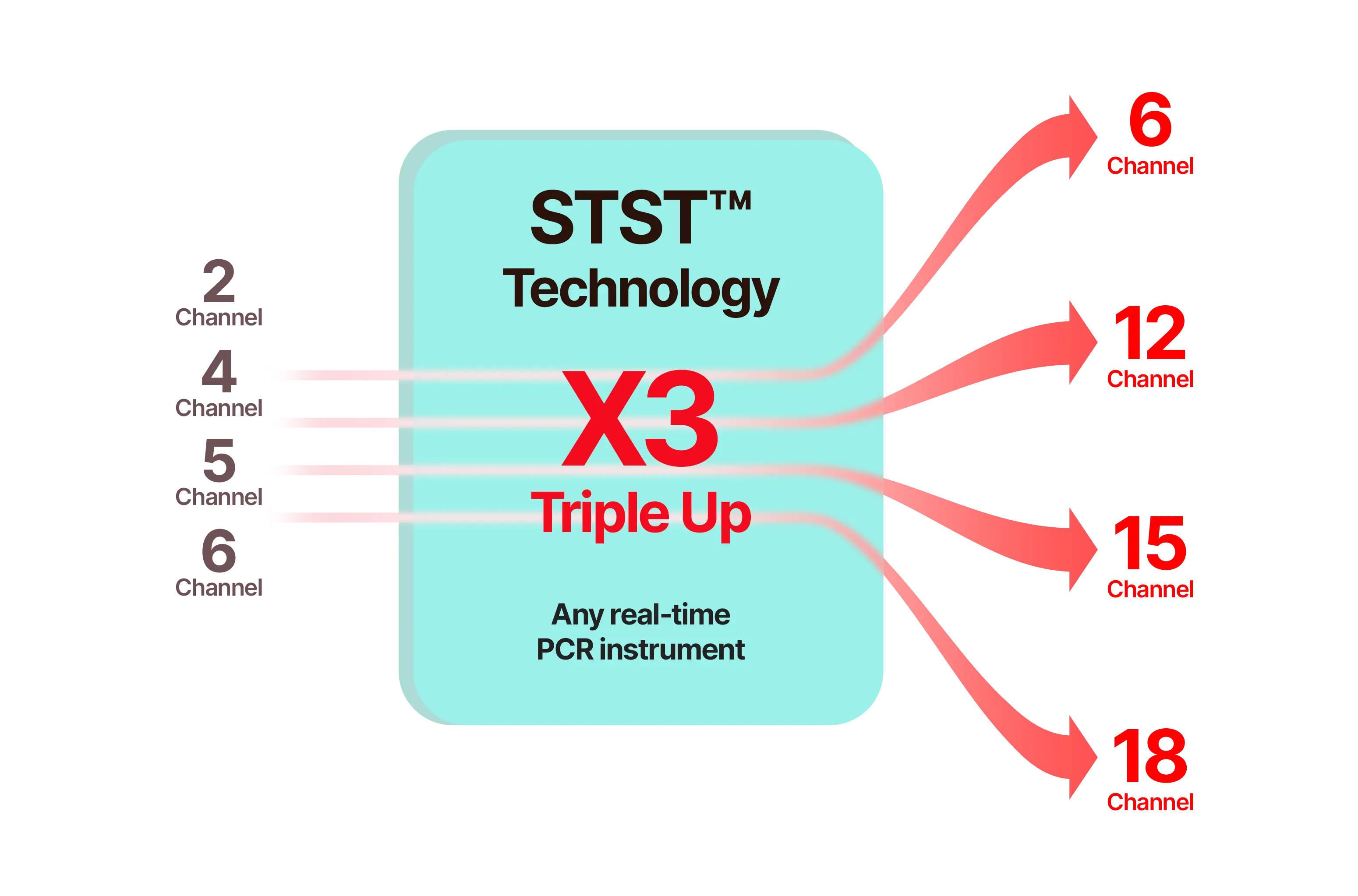
Expanded effective multiplexing capability
- Overcomes hardware limits to deliver enhanced multiplexing performance
- Enables multi-target quantification even on single-channel instruments
Faster decision-making with a simplified workflow
- Clear signal profiles allow rapid interpretation and reporting
- Reduced analysis time supports quicker clinical decisions